Copy Number Alterations (CNAs) - Central Premise
Copy-number alterations (CNAs) are changes in the allelic quantity of genes: from 2N copies to 1N, 0N (losses), or 3N to 200+N (gains). They are often formed through entire chromosome gains or losses. CNAs, which complement single-nucleotide variants and short insertion-deletions (often simply referred to as “mutations”), are the most common genetic alterations in cancer cells.
Improved therapies have been somewhat limited in cancers without kinase oncogene mutations. Some cancers, like ovarian cancer, triple-negative breast cancer, and serous uterine cancer have almost no canonical mutations which can be targeted by new therapies. However, these same cancers typically have exceptionally aberrant chromosomes. Patterns of chromosome losses and gain can be mapped to molecular pathways, indicating new cancer addictions and vulnerabilities. The lab assesses which gene copy-number alterations drive cancer biology, how they do so, and if targeting those alterations can lead to novel treatments.
Copy-number alterations (CNAs) are changes in the allelic quantity of genes: from 2N copies to 1N, 0N (losses), or 3N to 200+N (gains). They are often formed through entire chromosome gains or losses. CNAs, which complement single-nucleotide variants and short insertion-deletions (often simply referred to as “mutations”), are the most common genetic alterations in cancer cells.
Improved therapies have been somewhat limited in cancers without kinase oncogene mutations. Some cancers, like ovarian cancer, triple-negative breast cancer, and serous uterine cancer have almost no canonical mutations which can be targeted by new therapies. However, these same cancers typically have exceptionally aberrant chromosomes. Patterns of chromosome losses and gain can be mapped to molecular pathways, indicating new cancer addictions and vulnerabilities. The lab assesses which gene copy-number alterations drive cancer biology, how they do so, and if targeting those alterations can lead to novel treatments.
Cell Biology of Metallothioneins: A Unique Gene Cluster Influenced by Aneuploidy
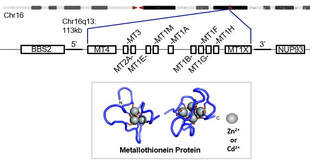
Background: While the autophagy pathway contains genes on many different chromosomes, some molecular pathways evolved from duplicated genes on a single chromosomal region. One such duplicated gene cluster is the metallothionein cluster. Metallothioneins are similar in function to hemoglobin in that their primary biochemical function is to chelate cations. Unlike hemoglobin, metallothioneins do not chelate iron, but rather zinc. Zinc is critical as a cofactor for many enzymes and transcription factors, including p53. In addition to zinc, metallothioneins also have high affinity to potentially toxic environmental and therapeutic metals: cadmium and platinum are prime examples. Much of the previous research performed on metallothioneins is correlative. Due to new tools, cell biology can be more precisely studied by carefully controlling metallothionein expression.
Current research: Metallothioneins are interesting to study from a genetic standpoint because all expressed, protein-coding genes are encoded on a single, relatively short region of a chromosome. In humans, these are 11 genes encoded sequentially on Chromosome 16. In mice, the 4 mouse metallothioneins are encoded sequentially on a syntenic region of Chromosome 8. This clustered arrangement allows us to test two important questions:
Unique techniques used in this area of research: Advanced genetic techniques are necessary to study how metallothioneins alter cancer cell biology. In addition, there are opportunities to screen for brand new drugs which target the effects of altered metallothionein expression.
Background: While the autophagy pathway contains genes on many different chromosomes, some molecular pathways evolved from duplicated genes on a single chromosomal region. One such duplicated gene cluster is the metallothionein cluster. Metallothioneins are similar in function to hemoglobin in that their primary biochemical function is to chelate cations. Unlike hemoglobin, metallothioneins do not chelate iron, but rather zinc. Zinc is critical as a cofactor for many enzymes and transcription factors, including p53. In addition to zinc, metallothioneins also have high affinity to potentially toxic environmental and therapeutic metals: cadmium and platinum are prime examples. Much of the previous research performed on metallothioneins is correlative. Due to new tools, cell biology can be more precisely studied by carefully controlling metallothionein expression.
Current research: Metallothioneins are interesting to study from a genetic standpoint because all expressed, protein-coding genes are encoded on a single, relatively short region of a chromosome. In humans, these are 11 genes encoded sequentially on Chromosome 16. In mice, the 4 mouse metallothioneins are encoded sequentially on a syntenic region of Chromosome 8. This clustered arrangement allows us to test two important questions:
- Do aneuploid losses of metallothioneins drive cancer biology?
- Do losses (or alternatively, over-expression) of metallothioneins create druggable cancer vulnerabilities?
Unique techniques used in this area of research: Advanced genetic techniques are necessary to study how metallothioneins alter cancer cell biology. In addition, there are opportunities to screen for brand new drugs which target the effects of altered metallothionein expression.
- CRISPR-Cas9 modulation of the gene cluster, including marker gene replacement
- Modified dCas9 fusion proteins which can function as synthetic transcription factors of the entire gene cluster
- Newly generated inducible mouse models of metallothionein expression, targeted to regions where cancer originates from
- Multiple-thousand drug candidate screening using high-content imaging and robotics
Turning Handicapped Autophagy into a New Cancer Therapy
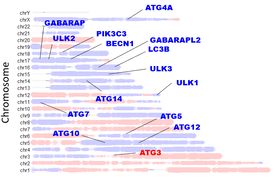
Background: Dr. Delaney previously established that autophagy genes are commonly on chromosome regions which are deleted in high-grade serous ovarian cancer, the deadliest gynecologic cancer resulting in over 10,000 deaths in the US annually. Autophagy is a cellular recycling system essential to maintain the energetics and proliferation of cancer cells. This finding of deletion patterns led to the hypothesis that reduced gene copy number prevented these ovarian cancer cells from being fully capable of increasing rates of autophagy in response to stress. This turned out to be the case; losses of genes mapped well to an impaired autophagosome-lysosome recycling system. Due to this vulnerable system, cancer cells could be targeted by therapeutics which either impair recycling further and/or create more molecular damage which autophagy is normally used to recycle. Normal cells are able to efficiently coordinate autophagy and clear this damage, but ovarian cancer cells cannot, and perish instead (video). We previously found targeting autophagy to be very promising in diminishing tumor burden in ovarian cancer mouse models.
Current research: It is currently unclear how to best target autophagy to lead to selective cancer cell death. We have found that a patient-data-centered approach, using clinical trial data on side effect, has led to very effective combinations. We term these combined autophagy drugs “Combination Of Autophagy Selective Therapeutics” or COAST. We are currently assessing:
Unique techniques used in this area of research: Translational research relies on excellent models used in the laboratory which can be recapitulate results we may find with real human patients. We use:
We are proud that our collaborations with medical doctors has resulted in an ongoing Phase I/II clinical trial first investigating the safety of up to 5-drug COAST in a human cancer patient setting (clinicaltrials.gov link). Some limited patient samples are available for biochemical research within the lab.
Background: Dr. Delaney previously established that autophagy genes are commonly on chromosome regions which are deleted in high-grade serous ovarian cancer, the deadliest gynecologic cancer resulting in over 10,000 deaths in the US annually. Autophagy is a cellular recycling system essential to maintain the energetics and proliferation of cancer cells. This finding of deletion patterns led to the hypothesis that reduced gene copy number prevented these ovarian cancer cells from being fully capable of increasing rates of autophagy in response to stress. This turned out to be the case; losses of genes mapped well to an impaired autophagosome-lysosome recycling system. Due to this vulnerable system, cancer cells could be targeted by therapeutics which either impair recycling further and/or create more molecular damage which autophagy is normally used to recycle. Normal cells are able to efficiently coordinate autophagy and clear this damage, but ovarian cancer cells cannot, and perish instead (video). We previously found targeting autophagy to be very promising in diminishing tumor burden in ovarian cancer mouse models.
Current research: It is currently unclear how to best target autophagy to lead to selective cancer cell death. We have found that a patient-data-centered approach, using clinical trial data on side effect, has led to very effective combinations. We term these combined autophagy drugs “Combination Of Autophagy Selective Therapeutics” or COAST. We are currently assessing:
- Potential side effects of COAST, particularly when combined with chemotherapy
- Sub-types of ovarian cancer which may best be treated using COAST
- Which other autophagy-disrupted cancer types may benefit from COAST
- Mechanism of synthetic lethality between COAST drugs
Unique techniques used in this area of research: Translational research relies on excellent models used in the laboratory which can be recapitulate results we may find with real human patients. We use:
- Genetically characterized cell lines which map well to human cancers
- Mouse models of ovarian cancer which retain an immune system
- Mouse models of human cancers using xenografts
- Spheroid models of ovarian cancer to mimic the adhesion-free ascites environment
- Whole-genome analysis of COAST-drug sensitivity modifiers (see section, below)
We are proud that our collaborations with medical doctors has resulted in an ongoing Phase I/II clinical trial first investigating the safety of up to 5-drug COAST in a human cancer patient setting (clinicaltrials.gov link). Some limited patient samples are available for biochemical research within the lab.
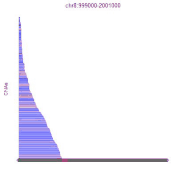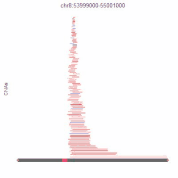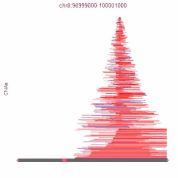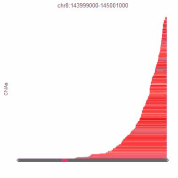
Bioinformatics of Copy-Number Alterations and Molecular Networks
Background: A major limitation to targeting mutant oncogenes and tumor suppressors is that many solid tumors are driven by rare mutations; for 10,000 patients, 10,000 drugs may have to be designed to fit these rare mutations. This is currently infeasible. High-grade serous ovarian cancer is mutant in p53 in all tumors, for example, but mutant p53 cannot currently be targeted to save lives in this disease. However, almost all solid tumors have common regions of chromosome gains or chromosome losses. If these can be targeted by drugs, each drug could be effective in double-digits percentages of patient, enabling feasibility of gene-targeted therapeutics. While our autophagy research (described above) serves as proof-of-principle that these chromosome aberrations can be targeted for improved therapies, much work remains to be done. We recently released an atlas of copy-number alterations which define tumor types, using advanced network analysis of biological pathways.
Importantly, the mathematical infrastructure developed for this cancer question is useful for any biological pathway: cancer related or not. Hence, we developed a suite of tools that molecular biologists can use for their genome-wide data. No programming expertise is required to use these tools, and analyses can be done on custom user data in as little as five clicks.
Current research:
Unique techniques used in this area of research:
Background: A major limitation to targeting mutant oncogenes and tumor suppressors is that many solid tumors are driven by rare mutations; for 10,000 patients, 10,000 drugs may have to be designed to fit these rare mutations. This is currently infeasible. High-grade serous ovarian cancer is mutant in p53 in all tumors, for example, but mutant p53 cannot currently be targeted to save lives in this disease. However, almost all solid tumors have common regions of chromosome gains or chromosome losses. If these can be targeted by drugs, each drug could be effective in double-digits percentages of patient, enabling feasibility of gene-targeted therapeutics. While our autophagy research (described above) serves as proof-of-principle that these chromosome aberrations can be targeted for improved therapies, much work remains to be done. We recently released an atlas of copy-number alterations which define tumor types, using advanced network analysis of biological pathways.
Importantly, the mathematical infrastructure developed for this cancer question is useful for any biological pathway: cancer related or not. Hence, we developed a suite of tools that molecular biologists can use for their genome-wide data. No programming expertise is required to use these tools, and analyses can be done on custom user data in as little as five clicks.
Current research:
- How can SWAN be utilized to discover novel biology within tumor subgroups?
- What are other best-case uses for SWAN in other diseases?
- What types of drugs are informed by SWAN analysis of chromosome alteration data?
- What new features should be administered in the online SWAN app?
- What new tools can and should be released to the community, focused on users with zero programming knowledge?
Unique techniques used in this area of research:
- R and Shiny App production
- Github, CRAN, and Bioconductor releases of bioinformatic R packages
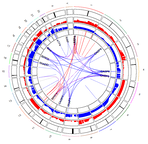
Origins of Copy-Number Alterations
Background: The scale of copy-number alterations in cancer is enormous, with the median tumor having 39% of its genome containing extra or less copies of genes. How does a pre-cancerous cell manage to develop so many changes that eventually enable them to become a full-blown metastatic tumor? Mutation of p53 is necessary but not sufficient for this level of genomic chaos.
Current research:
Unique techniques used in this area of research:
Background: The scale of copy-number alterations in cancer is enormous, with the median tumor having 39% of its genome containing extra or less copies of genes. How does a pre-cancerous cell manage to develop so many changes that eventually enable them to become a full-blown metastatic tumor? Mutation of p53 is necessary but not sufficient for this level of genomic chaos.
Current research:
- Do unique states of cancer cell biology, such as polyploid giant cancer cells and cancer stem cells, enable unusually high rates of chromosome aberrations?
- What mutations are necessary and/or sufficient to allow for surviving these highly chaotic cell divisions?
- Do repetitive elements play a major role in copy-number alteration biogenesis?
Unique techniques used in this area of research:
- Live-cell microscopy to observe unique cancer cell biology states (see video)
- Mouse models of spontaneous ovarian cancer development
- CRISPR-Cas9 screening of genes involved in these processes
Unbiased Genomics (All Projects)
Background: Genetics are the foundation to understanding biology. New techniques have enabled much deeper investigation of genetics relevant to disease. Our lab’s philosophy is that the best science is done when framed and executed in a manner minimizing bias. Fortunately, oncology is flush with experts utilizing the most advanced techniques known to science to enable genome scale analysis of complex biology.
Current research:
Unique techniques used in this area of research:
Background: Genetics are the foundation to understanding biology. New techniques have enabled much deeper investigation of genetics relevant to disease. Our lab’s philosophy is that the best science is done when framed and executed in a manner minimizing bias. Fortunately, oncology is flush with experts utilizing the most advanced techniques known to science to enable genome scale analysis of complex biology.
Current research:
- How can understanding RNA expression at the single-cell level inform how CNAs contribute to different aspects of the cancer microenvironment?
- Which genes are most relevant to drug sensitivity and resistance?
- How do gene and protein networks better inform biomarker patterns which may be of use clinically?
- How do our carefully manipulated genetic models contribute to genome-wide alterations?
Unique techniques used in this area of research:
- Next-generation sequencing of custom made libraries
- Single-cell sequencing
- Whole-genome Cas9 screens in specialized, perturbed biological systems
- Multi-omic bioinformatic analysis